Research
MAQC Consortium 2011–2014 (read more)
FDA SEQC, Nature Biotechnology (read more)
Bioinformatics (read more)
FEBS Journal (read more)
Gerontology (read more)
Impact of heavy tails in microarray analysis
Traditional microarray data analysis generally assumes Gaussian measurement noise – despite the sensitivity of the Normal Distribution to outliers. Especially for high-dimensional microarray data, Gaussian models are popular because of their computational efficiency in comparison to alternative approaches. We report on the first systematic study of the impact of noise model choice and its biological relevance.
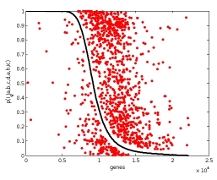 A hierarchical Bayesian model allows the principled
direct comparison of Gaussian models and robust
alternatives. Interestingly, heavy-tailed
distributions were the best fitting models for all the
examined data sets, spanning a wide range of
experiment types and measurement platforms. Moreover,
application of an appropriately heavy-tailed
t-distribution resulted in substantial changes
for differential expression analysis, strongly
affecting the functional categories
implicated. Traditional microarray analyses relying on
a Gaussian noise model thus not only distort
results for individual genes but yield biased
conclusions even at the higher level of functional
categories. In contrast, experimental evidence
strongly supports heavy tailed alternatives, and
different robust approaches agree well with one another.
A hierarchical Bayesian model allows the principled
direct comparison of Gaussian models and robust
alternatives. Interestingly, heavy-tailed
distributions were the best fitting models for all the
examined data sets, spanning a wide range of
experiment types and measurement platforms. Moreover,
application of an appropriately heavy-tailed
t-distribution resulted in substantial changes
for differential expression analysis, strongly
affecting the functional categories
implicated. Traditional microarray analyses relying on
a Gaussian noise model thus not only distort
results for individual genes but yield biased
conclusions even at the higher level of functional
categories. In contrast, experimental evidence
strongly supports heavy tailed alternatives, and
different robust approaches agree well with one another.
Reference
Posekany A, Felsenstein K, Sykacek P (2011) Assessing Robustness Issues in Microarray Data Analysis, Bioinformatics 27, 807-814. (read more | Supplement)
Return to group home or publications