Research
MAQC Consortium 2011–2014 (read more)
FDA SEQC, Nature Biotechnology (read more)
Bioinformatics (read more)
FEBS Journal (read more)
Gerontology (read more)
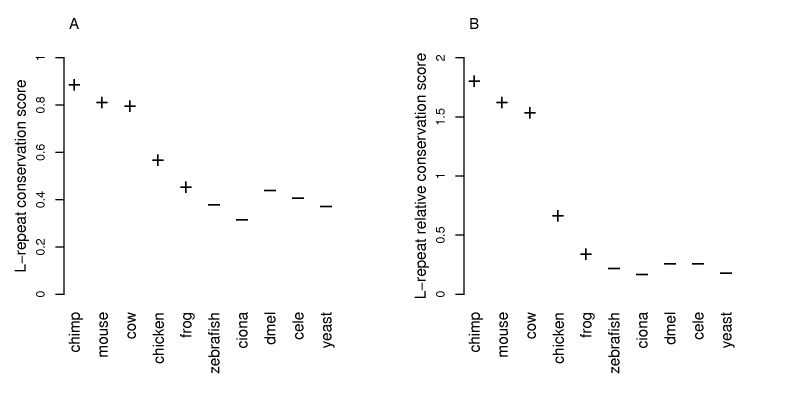
Figure 4: Sequence alignment based conservation scores. The left panel displays the average L-repeat conservation scores for the examined species, comprising five tetrapods ('+') and five non-tetrapods('-'). The right panel displays the average L-repeat relative conservation scores (see Methods). For the organisms closest to human, some signal peptides were identical. Perfectly matching signal pep-tides cannot provide information about the conservation of an L-repeat relative to the remaining signal sequence, as both show 100% sequence identity. Consequently, for tetrapods, scores excluding these proteins are shown. This Figure displays similarity based scores. Analogous scores based on identity% are shown in Suppl. Fig. S6.
(read more | Supplement | preprint)