
 Online Supplement for
Online Supplement forThis section provides figures illustrating the observed expression changes (both trends and deviations from the trends), links to fully annotated tables of affected genes, and tables showing trends by GOslim category. Please scroll down for Figures S3 and S4.
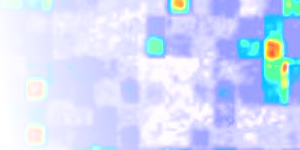
Figure 5 of our manuscript shows the systemic increase of expression for chromosome 5 genes in the trisomic plants, as a function of the average gene expression level (on the x-axis). Transcripts on chromosome 5 are coloured green, and the intensity dependent trend plus/minus standard deviation is plotted in magenta. The trend for transcripts on other chromosomes is shown in orange. The dashed vertical line marks the intensity A1+1 where the lower magenta and the upper orange lines cross and the trends are separated by 1+1 standard deviations. Surveys of trends thus focus on the strongly expressed transcripts to the right of the dashed lines, where the assay will be most accurate (*). The dotted vertical line indicates the lowest expression intensity for which a statistically significant change could be detected with p < 5% (Holm FWER). We here provide the figure in different resolutions [ low | medium | high ].
(*) The improved separation of more strongly expressed chromosome 5 genes from the trend of genes on other chromosomes that is evident in Figure 5 of our manuscript affects our ability to detect expression differences between trisomic and disomic samples. By plotting the percentage of genes on chromosome 5 that could be identified as having higher expression in trisomics, we can show that the sensitivity of our assay improves considerably for genes with an average intensity larger than approximately A1+1 and that the exact choice of the threshold makes little difference [ Athresh ]. To test the dosage-related increase in expression for genes on the trisomic chromosome 5 with lower expression levels, four moderately and five lowly expressed genes were selected for examination by high-sensitiviy qRT-PCR. Since the quantification of lowly expressed genes by real-time PCR can be non-trivial, genes were chosen with a well-documented exon/intron structure according to TAIR. The primers were designed to span the exon/intron junctions in order to ensure that PCR amplification did in fact report on differential gene expression rate and not, e.g., on residual genomic DNA. Consistent with the general chromosome 5 trend, a higher steady state transcript level in trisomics was indeed observed for the majority of these genes [ Fig. S1 and Fig. S2 ].
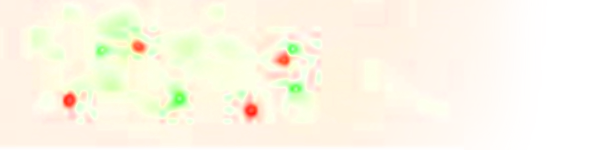
Figure 4 of our manuscript shows that all areas of the triplicated chromosome seem similarly affected [ cis.Mpos ] and that the observed trans effects appear to scatter randomly across chromosomes [ trans.Mpos ]. In these plots only strongly expressed genes, for which the effect could be assessed accurately are shown.
For a study of deviations from the cis chromosomal trend, this
trend was subtracted from the data and tests performed on the
zero-calibrated expression values. After successful calibration, the
magenta trend line traces the x-axis; and significant deviations
from the trend are shown as blue dots
[ cal0M: low |
medium |
high
resolution ].
We here also show the 100 genes most strongly deviating from the cis
trend in the context of an M(A) plot and their chromosomal
locations
[ chr5to0.MA |
chr5to0.Mpos ].
Also when all genes significantly differentially expressed relatively
to the chromosomal cis-trend are included in the figure
[ chr5to0.Mpos.plain ],
effects appear randomly distributed over the chromosome. Rainbow
colours indicate relative significance (red/yellow is highest,
blue/magenta is lowest).
Only a minority of genes is
below the general cis-trend and therefore dosage compensated or
down-regulated.
Figure S3
highlights in red the six epigenetic modifiers located on the
triplicated chromosome that are discussed in our manuscript (see
legend). All six fully follow the cis chromosomal trend of increased
expression.
Similarly, as an example of trans effects,
Figure S4
surveys differential regulation on chromosome 2. Red highlights
show the two epigenetic modifiers ROS1 and RDR5, prominently
up-regulated in the trisomic plants.
We considered two approaches for studying dosage compensation:
In summary, at least 13% of genes were dosage compensated, at least 3% of genes were fully dosage compensated, whereas at least 85% of genes were not dosage compensated. The manuscript text collects these summaries in a concise form, with test details given in the Methods section. Evidence from individual tests is quoted in parentheses in the main manuscript text.
These tables are TAB-delimited text files that can be loaded into any spreadsheet program. The largest tables are about 20MB in size. Subsets considering only strongly expressed genes are marked as bright. Calibrated cis data tested for deviation from the chromosomal trend is labelled Chr5to0.
| Group | A subset | p.adjust | Test subsets |
|---|---|
| Cis | bright | Holm FWER | All, Up, Down |
| Cis | all | Holm FWER | All, Up, Down |
| Cis | bright | B-Y FDR | All, Up, Down |
| Cis | all | B-Y FDR | All, Up, Down |
| Chr5to0 | bright | Holm FWER | All, Up, Down |
| Chr5to0 | all | Holm FWER | All, Up, Down, Diff ( = Up or Down ) |
| Chr5to0 | bright | B-Y FDR | All, Up, Down |
| Trans | bright | Holm FWER | All, Up, Down |
| Trans | all | Holm FWER | All, Up, Down, Diff ( = Up or Down ) |
The tables below are small TAB-delimited text files and can be loaded into any spreadsheet program or text editor. The first column shows the odds-ratio (OR) for the GOslim group being overrepresented in the test-set vs the entire chip, then follows a Holm adjusted FWER p-value, the relative (percentage) and absolute counts in test and reference sets, and the GOslim group being tested. Groups are sorted by significance of over- or underrepresentation in the test set. Only strongly expressed (bright) genes were considered, as our assay is most accurate for these.
| Group | Test subsets |
|---|---|
| Cis | Down, Not diff, Not up ( = Down or Not diff ) |
| Chr5to0 | Diff |
| Trans | Diff |