Research
MAQC Consortium 2011–2014 (read more)
FDA SEQC, Nature Biotechnology (read more)
Bioinformatics (read more)
FEBS Journal (read more)
Gerontology (read more)
While existing tools like limmaGUI make it easy to
rerun an existing model on new data, there is no support for
the definition of the model. Many efficient analyses follow a
balanced design and are based on an ANOVA. Defining a mixed
effects linear model and the relevant contrasts, however, is
 not trivial for biologists. In Sykcek et al. (2005) we
have introduced a new approach to facilitate the interaction
between experimentalists and data analysis experts for that task.
Processing is controlled by a simple text file that defines
and documents all steps of the analysis. The
Friendly
Statistics Package for Microarray Analysis provides the
tool as well as extensive instructions. In addition, we offer
hands-on lecture courses.
not trivial for biologists. In Sykcek et al. (2005) we
have introduced a new approach to facilitate the interaction
between experimentalists and data analysis experts for that task.
Processing is controlled by a simple text file that defines
and documents all steps of the analysis. The
Friendly
Statistics Package for Microarray Analysis provides the
tool as well as extensive instructions. In addition, we offer
hands-on lecture courses.
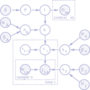 In Sykcek et al. (2007) we have introduced and
evaluated the power of a hierarchical Bayesian model for the
integrated analysis of several microarray data sets in order to
identify shared gene function. The approach has been been
tested in simulations as well as in the combined analysis of
in vitro and in vivo experiments using well
understood experimental conditions as a reference. As expected,
genes associated with apoptosis were identified as implicated
both in mouse mammary gland development and cell line growth factor
withdrawal. Our approach identified relevant genes with
higher sensitivity and specificity than traditional threshold
based alternatives.
In Sykcek et al. (2007) we have introduced and
evaluated the power of a hierarchical Bayesian model for the
integrated analysis of several microarray data sets in order to
identify shared gene function. The approach has been been
tested in simulations as well as in the combined analysis of
in vitro and in vivo experiments using well
understood experimental conditions as a reference. As expected,
genes associated with apoptosis were identified as implicated
both in mouse mammary gland development and cell line growth factor
withdrawal. Our approach identified relevant genes with
higher sensitivity and specificity than traditional threshold
based alternatives.
References
- Sykacek P, Furlong RA, Micklem G (2005) A friendly statistics package for microarray analysis. Bioinformatics 21, 4069–70. (read more)
- Sykacek P, Clarkson R, Print C, Furlong R, Micklem G (2007) Bayesian modelling of shared gene function. Bioinformatics 23, 1936–44. (read more | Supplement)
Return to group home or publications