Research
MAQC Consortium 2011–2014 (read more)
FDA SEQC, Nature Biotechnology (read more)
Bioinformatics (read more)
FEBS Journal (read more)
Gerontology (read more)
Characterization and improvement of RNA-Seq precision
Next generation sequencing is considered a promising novel technology also for the profiling of gene expression. We have performed a first systematic study of RNA-Seq reliability. Measurement precision determines the power of any analysis.
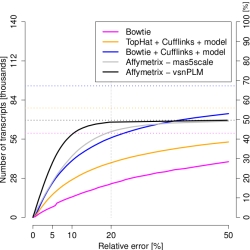 Expression estimates for the majority of spliceforms
were very noisy, with errors >20%. We have shown
that this is a direct consequence of the random
sampling of the highly skewed distribution of real
gene transcript abundances, independent of the
particular sequencing platform and protocol
employed. Most of the measurement power is spent on a
few strongly expressed transcripts, making the
remainder hard to measure. Each doubling of the read
depth adds <5% of reliably measured new
transcripts.
Expression estimates for the majority of spliceforms
were very noisy, with errors >20%. We have shown
that this is a direct consequence of the random
sampling of the highly skewed distribution of real
gene transcript abundances, independent of the
particular sequencing platform and protocol
employed. Most of the measurement power is spent on a
few strongly expressed transcripts, making the
remainder hard to measure. Each doubling of the read
depth adds <5% of reliably measured new
transcripts.
By exploiting gene models already at the alignment stage, we improved the number of reliably assessed spliceforms by 50% (blue curve). Yet standard microarrays performed 20% better (black curve). We therefore recommend and discuss iterative approaches for efficient expression profiling.
Reference
Łabaj PP, Leparc GG, Linggi BE, Markillie LM, Wiley HS, Kreil DP (2011) Characterization and improvement of RNA-Seq precision in quantitative transcript expression profiling, Bioinformatics 27, i383-i391. (read more | Supplement | reprint | ISMB video )
Return to group home or publications