Research
MAQC Consortium 2011–2014 (read more)
FDA SEQC, Nature Biotechnology (read more)
Bioinformatics (read more)
FEBS Journal (read more)
Gerontology (read more)
RNA interference in ageing research
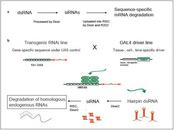 The search for genetic mechanisms affecting life-span and
ageing represents an important part of ageing research,
especially since the discovery of single-gene mutations with
dramatic effects on these traits. Considering its relative ease of
use, its power to specifically target arbitrary genes, and the
feasibility of genome-wide screens,
RNA interference (RNAi) has rapidly been adopted as a
technique for silencing gene expression. With afocus on the
model organisms Caenorhabditis elegans and
Drosophila melanogaster, and discussing recent
technical developments, this review highlights the current and
future impact of this technology in the field.
The search for genetic mechanisms affecting life-span and
ageing represents an important part of ageing research,
especially since the discovery of single-gene mutations with
dramatic effects on these traits. Considering its relative ease of
use, its power to specifically target arbitrary genes, and the
feasibility of genome-wide screens,
RNA interference (RNAi) has rapidly been adopted as a
technique for silencing gene expression. With afocus on the
model organisms Caenorhabditis elegans and
Drosophila melanogaster, and discussing recent
technical developments, this review highlights the current and
future impact of this technology in the field.
We show how RNAi has successfully been used to complement classic mutant studies. Moreover, we discuss the novel opportunities and challenges of an application of RNAi in genome-wide screens in D. melanogaster, which has become possible with the recent availability of a comprehensive transgenic RNAi library for the fly. We highlight, in particular, how the flexible control of RNAi induction can support the study of dynamic processes like ageing through specific experiments and the development of matching computational methods. In an overview of complementary approaches we discuss the challenge of extracting insight from the high-dimensional measurement datasets that are required for the study of dynamic effects and interaction dependencies.
Recent developments enable genome-wide screens with unprecedented temporal and spatial control of RNAi induction. Specific RNAi time-course experiments provide an opportunity for the analysis of high-resolution gene expression profiles capturing the dynamics of ageing-relevant processes and gene interactions. Research exploiting new avenues opened by the growing RNAi toolbox will considerably contribute to the next steps in researching the genetics of ageing and age-related diseases.
Reference
Minois N, Sykacek P, Godsey B, Kreil DP (2010) RNA Interference in Ageing Research. Gerontology, Epub ahead of print. (read more)
Return to group home or publications