Team
MAQC Consortium 2011–2014 (read more)
FDA SEQC, Nature Biotechnology (read more)
Bioinformatics (read more)
FEBS Journal (read more)
Gerontology (read more)
Pre-doctoral research fellow, bioinformatics
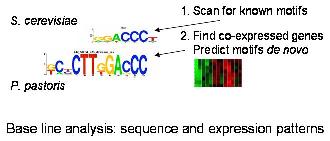
Joint modelling of gene expression patterns and regulatory potential of genome sequence regions for sensitive process identification
In this PhD project, the student will study gene expression patterns and examine the genomic sequence regions that may play a role in the regulation of gene expression. We will then explore established and novel approaches of looking at gene expression data and genomic sequence regions together. Such joint models have been shown to have superior power in the identification of biologically relevant patterns. We will use the recombinant expression of a payload protein and its relation to cell growth in the yeast Pichia pastoris as a test case. With our collaborators, we can test our predictions in the laboratory. (read more)
We are looking for a highly motivated post-graduate student with an interest in sequence analysis and expression profiling, in the context of studying transcriptional regulation. Familiarity with a warehouse / DBMS and data integration is an advantage. Experience with modern factor analysis (Bayesian PCA, ICA) is an advantage.
Please feel free to contact us with any questions about the type of work involved or the science behind the projects. If you are unsure about formal requirements or the application process, however, please ask the programme secretary, who will be happy to help.
Return to overview